3 (Or More) Ways to Open a CSV in Python
Ah. What a classic. The one piece of code that I end up writing over and over again, you would think I would have stashed it away by now. Not going to lie I usually have to Google it, while thinking, is this the right way? Should I just open the csv file and iterate it? Should I import the csv module? Should I just use Pandas? Does it matter? Probably not.
So, lets try them all. Not that it matters what’s slower, but sometimes you do run across the 2.5GB csv file, so it’s probably not a bad idea to check out the options.
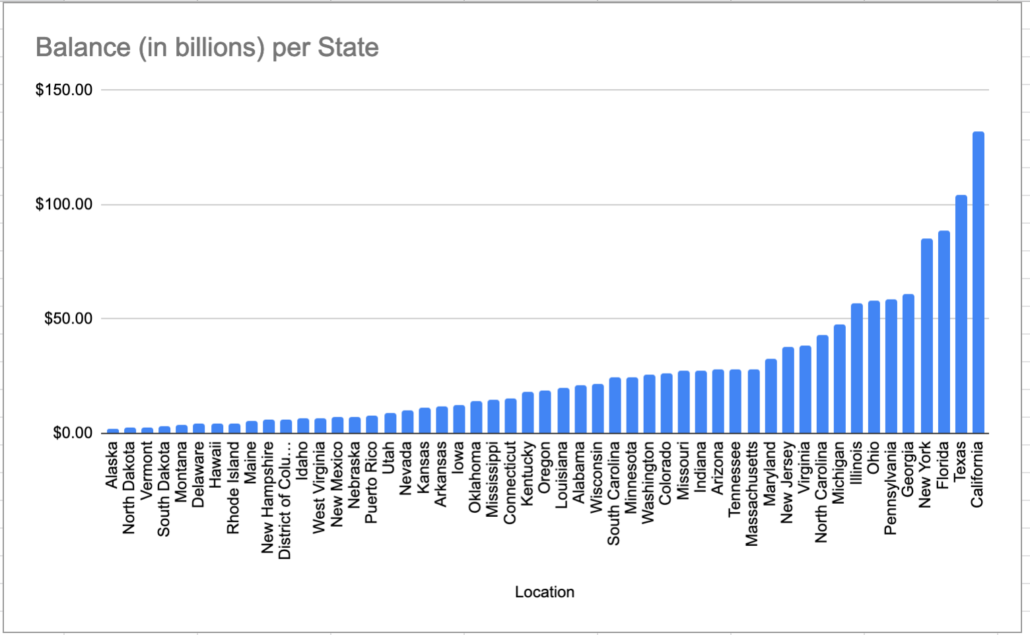
We will be using an open source data set, outstanding student load debt by state. All my code and the file can be found on GitHub. Let’s just open the file, read the rows and split the columns up and call that work.
Options for working with CSV files in Python.
- Just open it.
- Python standard csv module.
- Pandas.
Just Open the CSV File Already.
The first option is just to the open a file like you would anything else, and then read the lines one at at time. There are some sub options here.
- Read all the lines into list. readlines()
- Read one line at a time. readline()
- Read into a single string. read() – doesn’t apply in this situation for how we want to deal with data.
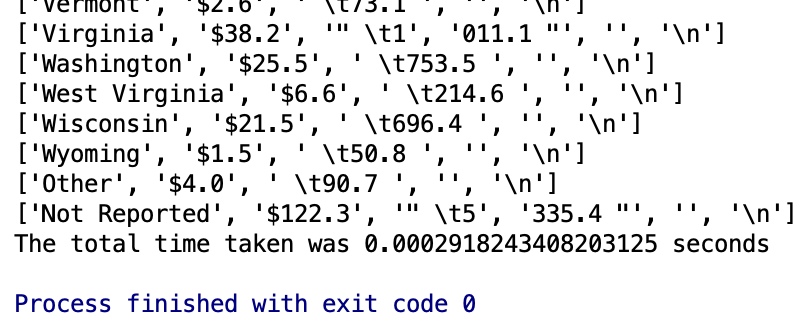
The idea behind just opening a file and calling readlines() or readline() is that it’s simple. With readlines() you will get a list back that contains a row for each line or record in your csv file. Using readline() you can just get one line at a time. Seems like you would maybe want to use readline() if you wanted to keep memory down, but most of the time who cares?
Let’s checkout readlines() first.
from time import time
def open_csv_file(file_location: str) -> object:
with open(file_location, 'r') as f:
data = f.readlines()
for line in data:
split_line(line)
def split_line(line: str) -> None:
column_data = line.split(',')
print(column_data)
if __name__ == '__main__':
t1 = time()
open_csv_file(file_location='PortfoliobyBorrowerLocation-Table 1.csv')
t2 = time()
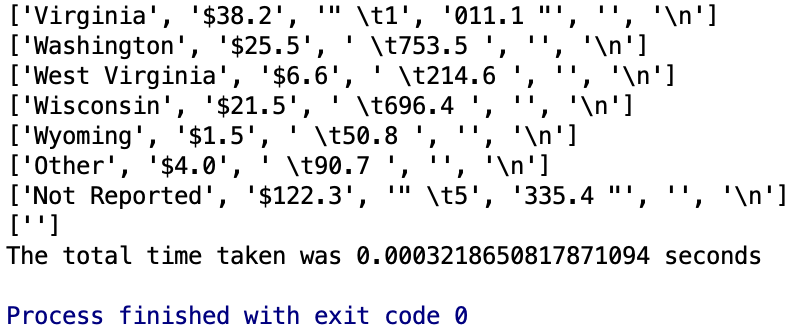
print('The total time taken was {t} seconds'.format(t=str(t2-t1)))Let’s try readline() next. We would expect it to be slightly lower because it just requires a touch more code.
from time import time
def open_csv_file(file_location: str) -> object:
with open(file_location, 'r') as f:
line = True
while line:
line = f.readline()
split_line(line)
def split_line(line: str) -> None:
column_data = line.split(',')
print(column_data)
if __name__ == '__main__':
t1 = time()
open_csv_file(file_location='PortfoliobyBorrowerLocation-Table 1.csv')
t2 = time()
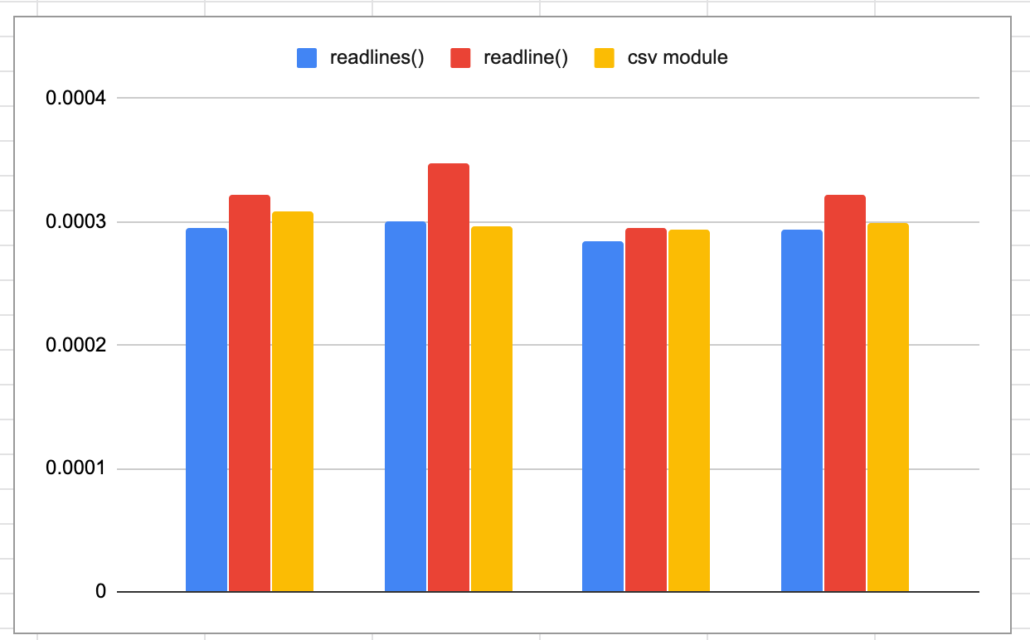
print('The total time taken was {t} seconds'.format(t=str(t2-t1)))I ran each method 3 times, below you can tell that readlines() is a little faster.
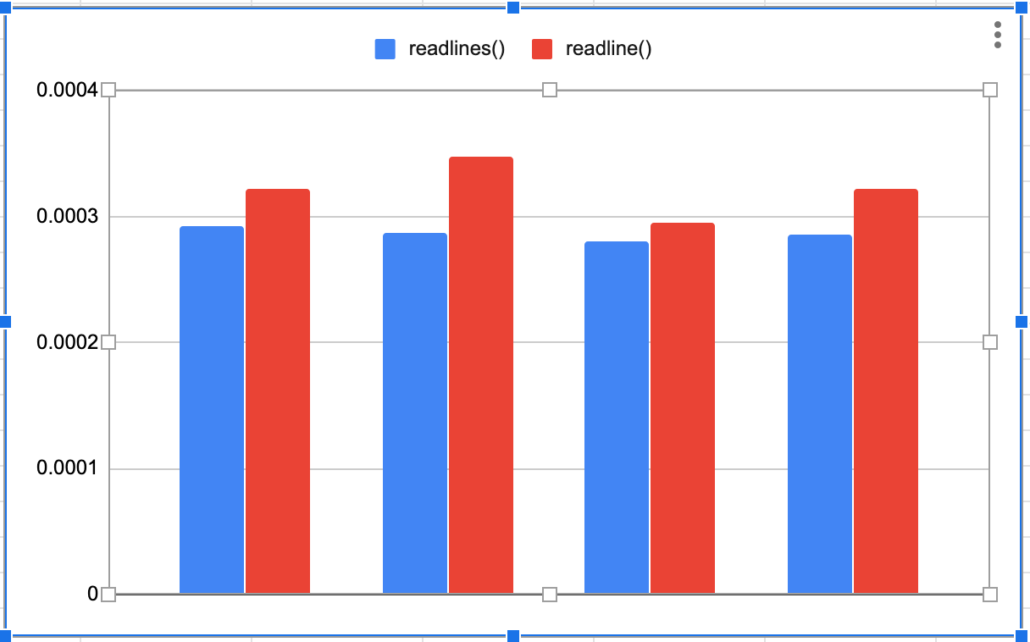
Import csv …. what could be easier?
Both those methods seem fairly straight forward. Let’s checkout the built in csv module in Python. This should be easier to use in theory because we won’t have to split out our own columns etc.
from time import time
import csv
def open_csv_file(file_location: str) -> object:
with open(file_location) as f:
csv_reader = csv.reader(f)
for row in csv_reader:
print(row)
if __name__ == '__main__':
t1 = time()
open_csv_file(file_location='PortfoliobyBorrowerLocation-Table 1.csv')
t2 = time()
print('The total time taken was {t} seconds'.format(t=str(t2-t1)))
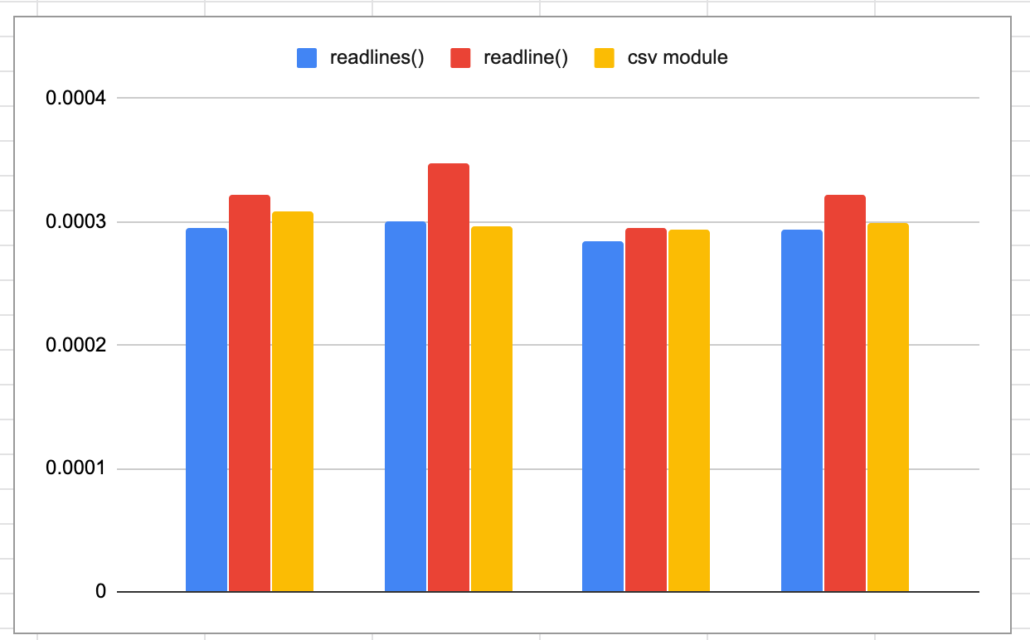
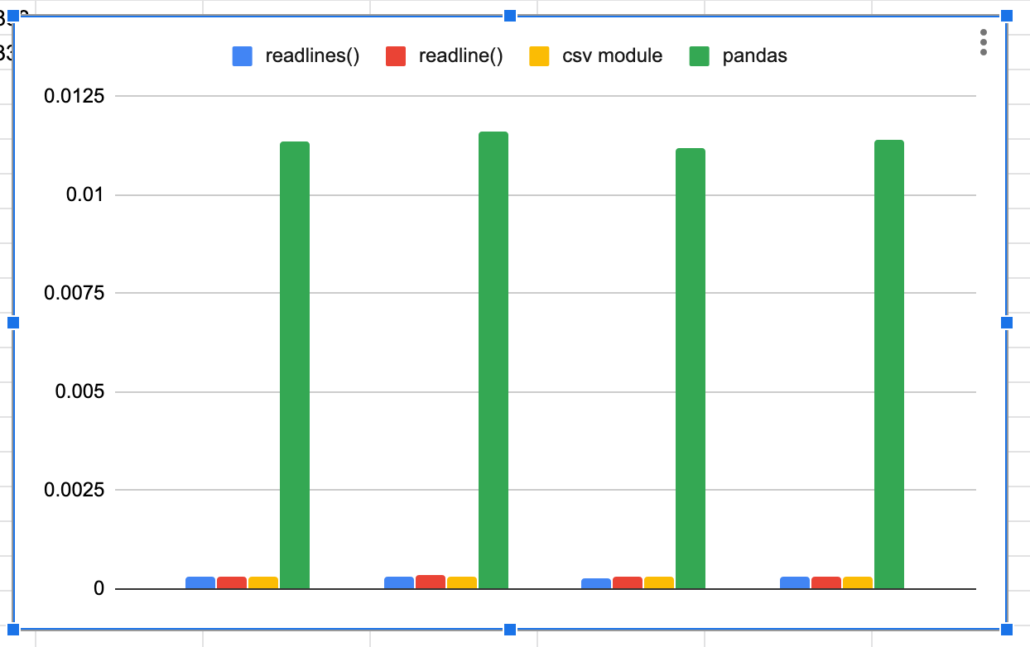
Interesting, faster then readline() but slightly slower then readlines() and splitting columns ourselves. This is a little strange to me, I just assumed that the csv module offered more then just convenience.
Who can say csv and Python in the same sentence and not think of Pandas? I have my complaints about it, but wit the rise of data science, it’s here to stay. I have to say, of all the options, reading a csv file with Pandas is the easiest to use and remember.
What makes Pandas nice is that to open a file into a dataframe all you have to do is call pandas.read_csv().
Also as you can see calling iterrows() will allow you to easily iterate over the rows.
from time import time
import pandas
def open_csv_file(file_location: str) -> object:
dataframe = pandas.read_csv(file_location)
for index, row in dataframe.iterrows():
print(row['Location'], row['Balance (in billions)'], row['Borrowers (in thousands)'])
if __name__ == '__main__':
t1 = time()
open_csv_file(file_location='PortfoliobyBorrowerLocation-Table 1.csv')
t2 = time()
print('The total time taken was {t} seconds'.format(t=str(t2-t1)))
Oh boy, easy to use by performance wise, yikes. Say goodbye to my nice looking chart!! HaHa!
Oh, and you can’t forget that piece of junk Dask. I know it wasn’t really made to read one csv file, but I have to poke at it anyways. If nothing else to make myself feel better.
from time import time
import dask.dataframe as dd
def open_csv_file(file_location: str) -> object:
df = dd.read_csv(file_location)
for index, row in df.iterrows():
print(row['Location'], row['Balance (in billions)'], row['Borrowers (in thousands)'])
if __name__ == '__main__':
t1 = time()
open_csv_file(file_location='PortfoliobyBorrowerLocation-Table 1.csv')
t2 = time()
print('The total time taken was {t} seconds'.format(t=str(t2-t1)))Bahaha!
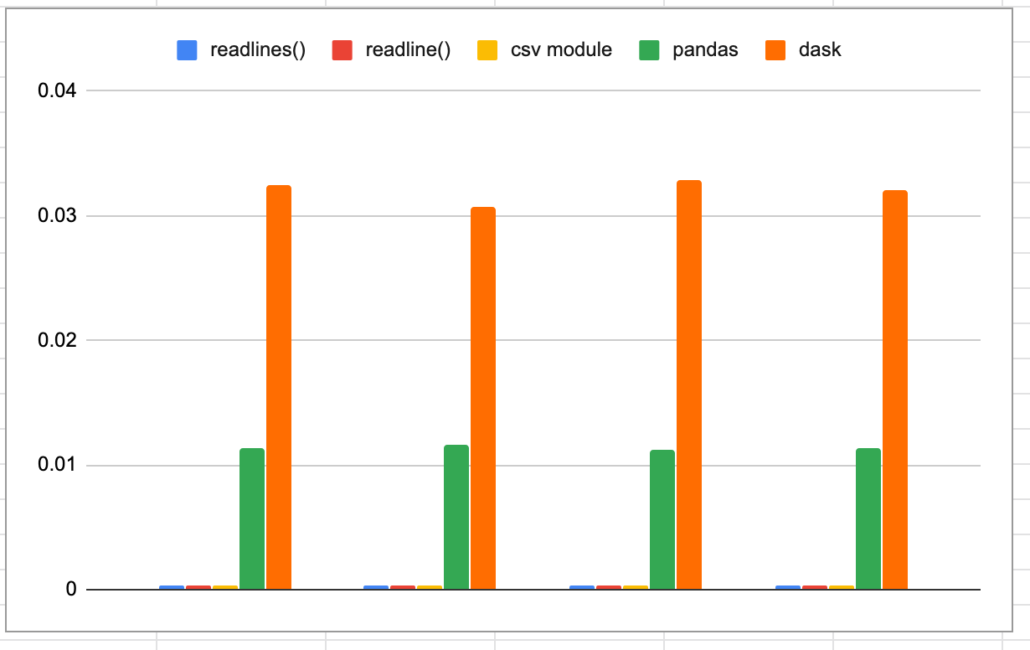
Nice! It’s always fun to go back to the simple stuff, loading csv files might be for the birds, but any data engineer is probably going to have to do it a few times a year. My vote is for readlines(), it’s fast and not that complicated.
I know some people might argue about the nuances of the different tools, and there are good reasons to use each one I’m sure. But, I think it’s important to just look at the basics of loading and iterating a CSV file with all the different tools. Mostly because in the real world we might just pick something in the heat of the moment, especially as a data engineer, and thousands of files later when things grow, come to the realization tool choice and speed did matter after all.
Oh by the way, in case you were curious and have heard a lot about the student loan debacle. You will notice we were using a data set of federal student loans per state. Here it is. Classic, way to go Cali.